Xenopus genome browser track hub
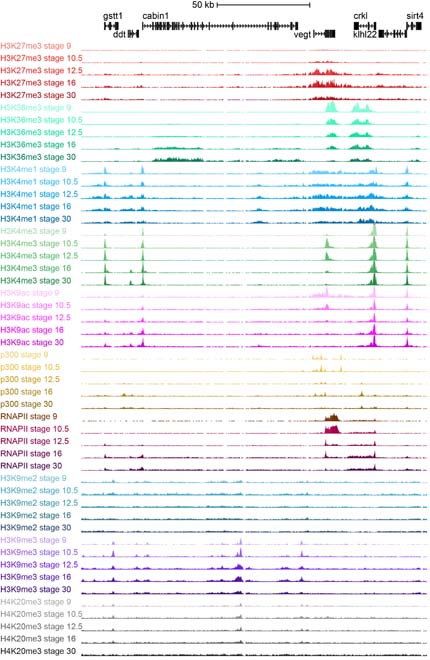
Epigenome reference maps
A track data hub has been generated providing access to Veenstra lab chromatin state maps, including ChIP-sequencing tracks of eight histone modifications, RNA polymerase II and the enhancer-binding protein p300 during Xenopus tropicalis development from blastula to larval stages.
Intergenic DNA contains functional genomic elements such as enhancers which are largely unannotated. The biochemical features of chromatin, such as post-translational modifications of histone proteins, determine its accessibility and function. The combination of different histone modifications can also be used to partition the genome in distinct chromatin states, such as promoters, enhancers, transcribed regions, and repressed heterochromatin.
The genome browser tracks have been generated for the histone modifications H3K4me1, H3K4me3, H3K9ac, H3K9me2, H3K9me3, H3K27me3, H3K36me3 and H4K20me3, in addition to RNA polymerase II and p300. At five stages of development (NF stages 9, 10.5, 12.5, 16 and 30) they have been mapped with or without repetitive reads, to allow assessing histone modfications at repetitive heterochromatic regions as well as unique regions within the genome.
(Click for larger image)
Updates
• X.tropicalis Epigenome reference maps published: Hontelez et al. (December 2015)
• X.tropicalis assembly v9 added to trackhub (February 2016)
• X.laevis assembly v9.1 and ChIP tracks added to trackhub (March 2016)
• X.tropicalis assembly v10.0 added to trackhub (March 2023)
Xenopus track hub instructions
The data can be browsed using a track data hub for the UCSC browser. Do the following to use this track hub:
- Visit the UCSC genome browser
- Click the button "track hubs"
- Select the tab "My Hubs"
- Copy-paste one of the following URL in the text box:
http://trackhub.science.ru.nl/hubs/veenstralab/trackhubs_public/xenopus/hub.txt
An older trackhub can also be used (no Xentro10_0):
http://trackhub.science.ru.nl/hubs/xenopus/hub.txt - Click "Add Hub"
- The newly added trackhub and its corresponding genome should be the default in the Genome Browser Gateway (home) page of the UCSC browser.
- If the trackhub is not selected, or to select the trackhub again among previously loaded trackhubs: In the genome browser, click the top menu item "Genomes" to go to the Genome Browser Gateway page. To the left, under "Represented species" the track hub "Xenopus UCSC hub" should be listed. Click it to select the trackhub.
- Select the Xenopus genome and assembly of interest under the drop-down under "Find Position", and click "Go" to move to a position in the genome.
- Browse the genome or modify track settings (right click on the track, or use track controls under main graphic)
- X. tropicalis v7.1, v8.0, v9.0 amd 10.0 with the Epigenome reference maps
- X. laevis v9.1
The generation of Epigenome reference maps has been supported by the US National Institutes of Health (NICHD, grant R01HD069344). The data can be freely used, please cite our papers when you do. Please contact Gert Jan Veenstra for other use. Published data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession numbers (listed with the publications below).
Genome data publications
Elurbe DM, Paranjpe SS, Georgiou G, van Kruijsbergen I, Bogdanovic O, Gibeaux R, Heald R, Lister R, Huynen MA, van Heeringen SJ, Veenstra GJC. 2017. Regulatory remodeling in the allo-tetraploid frog Xenopus laevis. Genome Biol., 18, page 198 (doi: 10.1186/s13059-017-1335-7.).
![]() Genome Biology
Genome Biology
![]() PDF
PDF
![]() NCBI GSE76059,
GSE92382,
GSE90898,
GSE67974,
GSE56586
NCBI GSE76059,
GSE92382,
GSE90898,
GSE67974,
GSE56586
![]()
Hontelez S., I. van Kruijsbergen, G. Georgiou, S. J. van Heeringen, O. Bogdanovic, R. Lister, G.J.C. Veenstra. 2015. Embryonic transcription is controlled by maternally defined chromatin state. Nature Commun., 6, 10148 (doi: 10.1038/ncomms10148).
![]() Nature Communications
Nature Communications
![]() PDF
PDF
![]() NCBI GSE67974
NCBI GSE67974
![]()
Heeringen, S.J. van, R.C. Akkers, I. van Kruijsbergen, M.A. Arif, L.L.P. Hanssen, N. Sharifi, G.J.C. Veenstra. 2014. Principles of nucleation of H3K27 methylation during embryonic development. Genome Research, 24, 401-410 (doi: 10.1101/gr.159608.113).
![]() Genome Research
Genome Research
![]() PDF
PDF
![]() NCBI GSE41161
NCBI GSE41161
![]()
Paranjpe SS, U.G. Jacobi, S.J. van Heeringen, G.J.C. Veenstra. 2013. A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics, 14,762 (doi:10.1186/1471-2164-14-762).
![]() BMC Genomics
BMC Genomics
![]() NCBI GSE43652
NCBI GSE43652
![]()
Bogdanovic, O., S.W. Long, S. J. van Heeringen, A.B. Brinkman, J.L. Gómez-Skarmeta, H.G. Stunnenberg, P.L. Jones and G.J.C. Veenstra. 2011. Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Research, 21, 1313-1327 (doi: 10.1101/gr.114843.110).
![]() Genome Research Online
Genome Research Online
![]()
![]() NCBI GSE23913
NCBI GSE23913
![]()
Van Heeringen, S.J., W. Akhtar, U.G. Jacobi, R.C. Akkers, Y. Suzuki, G.J.C. Veenstra. 2011. Nucleotide composition-linked divergence of vertebrate core promoter architecture. Genome Research, 21, 410-421.
![]() Genome Research Online
Genome Research Online
![]()
![]() NCBI GSE21482
NCBI GSE21482
![]()
Akkers, R.C., S.J. van Heeringen, U.G. Jacobi, E.M. Janssen-Megens, K.J. Françoijs, H.G. Stunnenberg and G.J.C. Veenstra. 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Developmental Cell, 17, 425-434.
![]() Dev.Cell Online
Dev.Cell Online
![]()
![]() PubMed Central
PubMed Central
![]() NCBI GSE14025
NCBI GSE14025
![]()
